Zvonimir Dogic talked about dynamic phenomena – things that move and change while consuming energy. Microtubules and microfilaments are useful systems to study… they can be observed directly. Kinesin doing some interesting sliding dynamics. I like that his movies have subtle timestamps in the upper left. His microscopy is so clean. I want to know what scope he was using and how it is maintained. He showed 1mm sized views with beautiful across-the-board quality. The focus stays perfect for a highly extended time. The depletion interaction – PEG exclusion – is a nice way to exclude things to the surface, for instance.
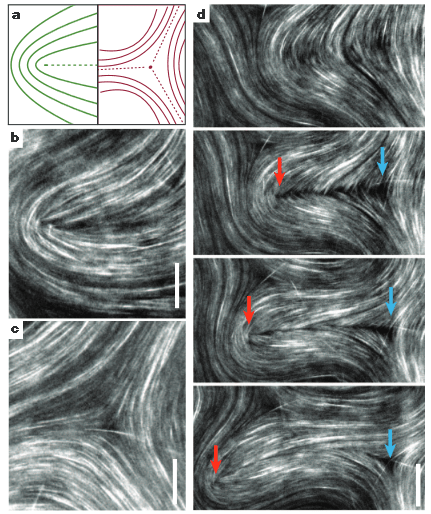 From “Spontaneous motion in hierarchically assembled active matter,” Tim Sanchez, Daniel T. N. Chen, Stephen J. DeCamp, Michael Heymann and Zvonimir Dogic, Nature 491, 431-434 (2012). Scale bars are 20 um.
From “Spontaneous motion in hierarchically assembled active matter,” Tim Sanchez, Daniel T. N. Chen, Stephen J. DeCamp, Michael Heymann and Zvonimir Dogic, Nature 491, 431-434 (2012). Scale bars are 20 um.
He showed great movies of microtubule bundles at interfaces forming an actively advexing chaotic gel. Capillary flow actually emerges from the system. The topic turned to the topology of liquid crystal defects which is a surprisingly universal aspect of mathematics.
Jonathan Doye talked about mechanical properties of DNA as modeled in a medium-grain simulation. The model is a simplified version of DNA that is still being improved to incorporate the finer features of DNA structure. But the coarse features – pairing, stiffness, etc. – are still useful. Including modeling origami and nanodevices like a walker. But one reasonable model conclusion was that mismatches can accelerate the rearrangement to the minimum. It looks lime this model might explain the some strange phenomena talked about at DNA19 by Niranjan Srinivas that showed an anomalously slow kinetic step in the strand displacement reaction.
Damien Woods talked about self-replicating nanostructures. He wanted to make a soup of DNA that could form into a long tubule, but would do so incredibly slowly unless there were little tubule seeds floating around. He then introduced some seeds; tubules grew. Even better, as they grew really big they could be made to break in half and thus create more tubes.
Carlos Castro showed some very dynamic 3D DNA origami structures which were very pretty. The assembled TEM movies make it seem very tangible. The structures could in principle be used to do some force spectroscopy by exploiting the entropic spring properties of ssDNA.
I was very happy to see Niranjan Srinivas from the Winfree lab give his talk on the kinetics of strand displacement. I have been trying to remember the details since DNA19. There are some anomalies in the rates of certain steps of toehold mediated strand displacement that are very counterintuitive. I gather that the issue is some subtle rearrangement that is captured by the models shown in Jonathan Doye’s talk.
Katherine Willets and Joel Harris talked about analyzing surface bound fluorescent DNA which was very informative. Making dyes that can be turned off is obviously very important. Inverse DNA-PAINT with temporary quenchers might be an interesting solution to some of the high-density issues that prevent good localization of the Gaussian centroid.
Ibuki Kawamata talked about some DNA crosslinked gels and showed some interesting results where a steel ball would be suspended on top of a gel, giving a quantitative readout of its degree of crosslinking. Could one analyze a movie of a steel ball falling through a gel (or use a magnetic sensor?) and compute the viscosity? That would be a pretty convenient viscometer.

You must be logged in to post a comment.